ADM1F SRT: single tank and two-phase anaerobic dynamic membrane bioreactor¶
This script is used to simulate a single tank suspended anaerobic dynamic membrane digester and a novel two-phase anaerobic dynamic membrane bioreator with separated SRT and HRT. In the two-phase reactor the effluent (model output) from the first phase dynamic membrane bioreactor is converted to the influent (model input) for the second-phase anaerobic dynamic membrane bioreactor as shown in the figure.
Note: Before running the ADM1F SRT simulations see intructions on how to compile adm1f_srt.cxx in the User Guide.
Mass balance equation with SRT:
The ADM1 and ADM1F solve the mass balance equation (i.e. mass_change = mass_in – mass_out + reaction). ADM1F_SRT version of the model includes solid retention time (\(t_{res,X}\)) as shown in the equation below.
Authors: Wenjuan Zhang, Elchin Jafarov, Kuang Zhu
[1]:
# Load packages
import numpy as np
import pandas as pd
import os
import seaborn as sns
import adm1f_utils as adm1fu
import matplotlib.pyplot as plt
%matplotlib inline
[2]:
# navigate to simulations folder
os.chdir('../simulations')
[3]:
# Grab the names and unit of all the outputs
(output_name,output_unit)=adm1fu.get_output_names()
[4]:
#check the path to the executable
!echo $ADM1F_EXE
/Users/elchin/project/ADM1F_WM/build/adm1f
Single tank anaerobic dynamic membrane bioreactor (AnDMBR)¶
To simulate the single tank AnDMBR that has separated SRT and HRT, We can call the functionreactor1 with different Q (flow-rate), Vliq (reactor volume), t_resx (SRT-HRT) values, the function will return the corresponding output
Usage example: reactor1(Q=100, t_resx=30, Vliq=300)
(Unit) Q: [m3/d], t_resx: [day], Vliq: [m3]
[5]:
# testing different SRTs=[0,1,2,...9] on the one-phase reactor
resx_list = [i for i in range(10)]
# setup the matrix with columns correspoding SRTs and rows to outputs
output1_resx = np.zeros((len(resx_list), 67))
# here we utilize back euler solver and adataptive time step
# for more command options see "User Guide/Running ADM1F/step 5"
options='-ts_type beuler -ts_adapt_type basic -ts_max_snes_failures -1 -steady'
for i in range(len(resx_list)):
output1_resx[i] = adm1fu.reactor1(opt=options, Vliq=300, Q=600, t_resx=resx_list[i])
np.savetxt('output_1phase.csv',output1_resx,delimiter=',',fmt='%1.4e')
Reactor run, phase-one:
$ADM1F_EXE -ts_type beuler -ts_adapt_type basic -ts_max_snes_failures -1 -steady -Vliq 300 -t_resx 0 -influent_file influent_cur.dat
indicator-228.out
Reactor run, phase-one:
$ADM1F_EXE -ts_type beuler -ts_adapt_type basic -ts_max_snes_failures -1 -steady -Vliq 300 -t_resx 1 -influent_file influent_cur.dat
indicator-307.out
Reactor run, phase-one:
$ADM1F_EXE -ts_type beuler -ts_adapt_type basic -ts_max_snes_failures -1 -steady -Vliq 300 -t_resx 2 -influent_file influent_cur.dat
indicator-437.out
Reactor run, phase-one:
$ADM1F_EXE -ts_type beuler -ts_adapt_type basic -ts_max_snes_failures -1 -steady -Vliq 300 -t_resx 3 -influent_file influent_cur.dat
indicator-563.out
Reactor run, phase-one:
$ADM1F_EXE -ts_type beuler -ts_adapt_type basic -ts_max_snes_failures -1 -steady -Vliq 300 -t_resx 4 -influent_file influent_cur.dat
indicator-521.out
Reactor run, phase-one:
$ADM1F_EXE -ts_type beuler -ts_adapt_type basic -ts_max_snes_failures -1 -steady -Vliq 300 -t_resx 5 -influent_file influent_cur.dat
indicator-516.out
Reactor run, phase-one:
$ADM1F_EXE -ts_type beuler -ts_adapt_type basic -ts_max_snes_failures -1 -steady -Vliq 300 -t_resx 6 -influent_file influent_cur.dat
indicator-534.out
Reactor run, phase-one:
$ADM1F_EXE -ts_type beuler -ts_adapt_type basic -ts_max_snes_failures -1 -steady -Vliq 300 -t_resx 7 -influent_file influent_cur.dat
indicator-542.out
Reactor run, phase-one:
$ADM1F_EXE -ts_type beuler -ts_adapt_type basic -ts_max_snes_failures -1 -steady -Vliq 300 -t_resx 8 -influent_file influent_cur.dat
indicator-556.out
Reactor run, phase-one:
$ADM1F_EXE -ts_type beuler -ts_adapt_type basic -ts_max_snes_failures -1 -steady -Vliq 300 -t_resx 9 -influent_file influent_cur.dat
indicator-576.out
[6]:
df1_resx = pd.read_csv('output_1phase.csv', sep=',', header=None)
df1_resx.columns = output_name
df1_resx.insert(0,"T_resx",resx_list)
Relation between t_resx and output when Vliq=300m3, Q=600m3/d
[7]:
# check the results with increasing T_resx, we should expect decrease in Ssu
df1_resx
[7]:
| T_resx | Ssu | Saa | Sfa | Sva | Sbu | Spro | Sac | Sh2 | Sch4 | ... | Alk | NH3 | NH4 | LCFA | percentch4 | energych4 | efficiency | VFA/ALK | ACN | sampleT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 2152.800 | 324.2700 | 7382.70 | 2381.700 | 3667.300 | 2542.700 | 8263.5 | 25.643000 | -8.963700e-82 | ... | 44957.0 | 0.053067 | 1074.00 | 7382.70 | -1.133000e-81 | 83.808 | 3.9774 | 0.351490 | -3.203400e-72 | 0.5 |
| 1 | 1 | 123.060 | 49.1670 | 7751.20 | 2574.400 | 4202.900 | 3244.500 | 9600.7 | 30.352000 | 3.859800e-12 | ... | 59182.0 | 0.047701 | 1135.60 | 7751.20 | 1.599000e-12 | 100.630 | -76.4800 | 0.310840 | 1.255000e-08 | 0.5 |
| 2 | 2 | 63.057 | 26.6380 | 7839.00 | 2610.000 | 4268.600 | 3305.000 | 9742.9 | 30.772000 | 1.085400e-09 | ... | 61421.0 | 0.048208 | 1150.90 | 7839.00 | 4.422100e-10 | 102.210 | -155.0200 | 0.304150 | 2.777100e-07 | 0.5 |
| 3 | 3 | 39.313 | 17.3450 | 908.19 | 41.129 | 54.118 | 53.411 | 3339.7 | 0.000662 | 2.574900e+02 | ... | 16289.0 | 2.513100 | 963.03 | 908.19 | 5.579800e+01 | 209.370 | -218.4400 | 0.200760 | 9.097200e+00 | 0.5 |
| 4 | 4 | 30.634 | 13.5750 | 457.20 | 30.592 | 39.989 | 35.895 | 3066.6 | 0.000515 | 2.653300e+02 | ... | 18846.0 | 3.157300 | 963.70 | 457.20 | 5.623900e+01 | 214.510 | -304.2100 | 0.157850 | 1.324200e+01 | 0.5 |
| 5 | 5 | 25.261 | 11.2240 | 309.54 | 24.543 | 31.958 | 27.254 | 2856.4 | 0.000424 | 2.695900e+02 | ... | 21319.0 | 3.693200 | 966.73 | 309.54 | 5.648800e+01 | 217.230 | -390.0200 | 0.129290 | 1.778600e+01 | 0.5 |
| 6 | 6 | 21.606 | 9.6181 | 236.08 | 20.615 | 26.775 | 22.111 | 2685.8 | 0.000362 | 2.726300e+02 | ... | 23721.0 | 4.147900 | 970.17 | 236.08 | 5.666900e+01 | 219.140 | -475.6200 | 0.108900 | 2.279000e+01 | 0.5 |
| 7 | 7 | 18.960 | 8.4513 | 192.14 | 17.859 | 23.153 | 18.702 | 2543.7 | 0.000317 | 2.750200e+02 | ... | 26050.0 | 4.540600 | 973.69 | 192.14 | 5.681200e+01 | 220.630 | -560.8900 | 0.093693 | 2.825000e+01 | 0.5 |
| 8 | 8 | 16.955 | 7.5652 | 162.87 | 15.818 | 20.479 | 16.276 | 2423.2 | 0.000284 | 2.770000e+02 | ... | 28311.0 | 4.885500 | 977.21 | 162.87 | 5.692900e+01 | 221.870 | -645.8400 | 0.081984 | 3.416600e+01 | 0.5 |
| 9 | 9 | 15.383 | 6.8691 | 141.96 | 14.244 | 18.422 | 14.462 | 2319.3 | 0.000257 | 2.786900e+02 | ... | 30505.0 | 5.192300 | 980.68 | 141.96 | 5.702800e+01 | 222.920 | -730.4900 | 0.072728 | 4.053400e+01 | 0.5 |
10 rows × 68 columns
[8]:
# correlation between SRT and output
plt.figure(figsize=(24,1))
corr1=df1_resx.corr()
sns.heatmap(corr1.iloc[0:1,-67:], xticklabels=df1_resx.columns[-67:], yticklabels=df1_resx.columns[0:1], cmap=sns.diverging_palette(220, 10, as_cmap=True))
plt.title('Correlation Matrix [t_resx/results]',fontsize=20);

Configurations
Configuration |
Vliq (m\(^3\)) |
t_resx (d) |
Q (m\(^3\)/d) |
|---|---|---|---|
Default |
3400 |
0 |
134 |
Phase 1 |
340 |
1.5 |
618 |
Phase 2 |
3400 |
700 |
618/— |
where t_resx = SRT - HRT
[9]:
config_default = {'Vliq':3400, 't_resx':0, 'Q':134}
config1 = {'Vliq':340, 't_resx':1.5, 'Q':618}
config2 = {'Vliq':3400, 't_resx':700, 'Q':618}
Default Configuration¶
[10]:
# output using default configuration
ls_default = adm1fu.reactor1(**config_default).tolist()
df_default = pd.DataFrame(data = [ls_default],columns=output_name, index=['Default Configuration'])
df_default
Reactor run, phase-one:
$ADM1F_EXE -Vliq 3400 -t_resx 0 -influent_file influent_cur.dat
indicator-034.out
[10]:
| Ssu | Saa | Sfa | Sva | Sbu | Spro | Sac | Sh2 | Sch4 | Sic | ... | Alk | NH3 | NH4 | LCFA | percentch4 | energych4 | efficiency | VFA/ALK | ACN | sampleT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Default Configuration | 7.17674 | 3.21796 | 54.9446 | 6.39778 | 8.23228 | 6.08339 | 1963.6 | 0.00012 | 48.3303 | 639.764 | ... | 8438.52 | 9.32395 | 1026.69 | 54.9446 | 56.905 | 65.228 | 53.3712 | 0.220453 | 109.164 | 25.3731 |
1 rows × 67 columns
Configuration 1¶
[11]:
# output using configuration 1
ls_config1 = adm1fu.reactor1(**config1).tolist()
df_config1 = pd.DataFrame(data = [ls_config1],columns=output_name, index=['Phase 1 Configuration'])
df_config1
Reactor run, phase-one:
$ADM1F_EXE -Vliq 340 -t_resx 1.5 -influent_file influent_cur.dat
indicator-051.out
[11]:
| Ssu | Saa | Sfa | Sva | Sbu | Spro | Sac | Sh2 | Sch4 | Sic | ... | Alk | NH3 | NH4 | LCFA | percentch4 | energych4 | efficiency | VFA/ALK | ACN | sampleT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phase 1 Configuration | 80.5616 | 33.4704 | 7808.75 | 2598.14 | 4246.87 | 3285.25 | 9696.05 | 28.5069 | 7.737870e-30 | 147.214 | ... | 60316.4 | 0.048011 | 1145.6 | 7808.75 | 3.164870e-30 | 104.203 | -104.737 | 0.308161 | 1.202840e-17 | 0.550162 |
1 rows × 67 columns
Configuration 2¶
[12]:
# output using configuration 2
ls_config2 = adm1fu.reactor1(**config2).tolist()
df_config2 = pd.DataFrame(data = [ls_config2],columns=output_name, index=['Phase 2 Configuration'])
df_config2
Reactor run, phase-one:
$ADM1F_EXE -Vliq 3400 -t_resx 700 -influent_file influent_cur.dat
indicator-025.out
[12]:
| Ssu | Saa | Sfa | Sva | Sbu | Spro | Sac | Sh2 | Sch4 | Sic | ... | Alk | NH3 | NH4 | LCFA | percentch4 | energych4 | efficiency | VFA/ALK | ACN | sampleT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phase 2 Configuration | 2.70068 | 1.20723 | 19.0168 | 2.34856 | 3.01652 | 2.16597 | 1125.18 | 0.000045 | 75.1774 | 952.376 | ... | 21574.7 | 14.7549 | 1186.64 | 19.0168 | 58.194 | 255.569 | -899.833 | 0.049221 | 3302.96 | 5.50162 |
1 rows × 67 columns
Two-Phase anaerobic dynamic membrane bioreactor¶
To simulate a novel two-phase AnDMBR, We can call the function reactor2 with different Q1 (phase 1 flow rate), Q2 (phase 2 flow rate, normally equal to Q1), Vliq1 (phase 1 volume), Vliq2 (phase 2 volume), t_resx1 (SRT-HRT for phase 1), and t_resx2 (SRT-HRT for phase 1) values. In this function, the model output from phase 1 will be extracted and converted to the model input for phase 2. The function will return phase1 output and phase2 output
Usage example: reactor2(Q1=100, t_resx1=30, t_resx2=100, Vliq1=300, Vliq2=3000)
[13]:
from IPython.display import Image
print ('Schematics of the two-phase reactor')
Image(filename='../notebooks/2phase_reactor.png')
Schematics of the two-phase reactor
[13]:
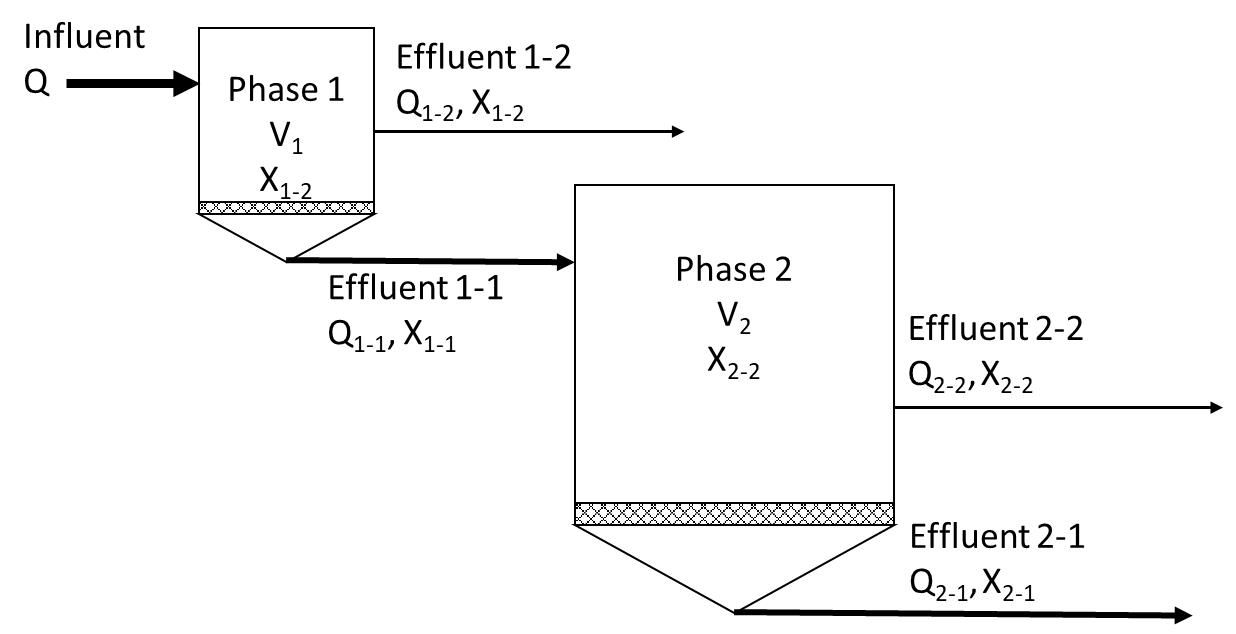
Config12¶
Configurations
Configuration |
Vliq (m\(^3\)) |
t_resx (d) |
Q (m\(^3\)/d) |
|---|---|---|---|
Default |
3400 |
0 |
134 |
Phase 1 |
340 |
1.5 |
618 |
Phase 2 |
3400 |
700 |
618/— |
where t_resx = SRT - HRT
[14]:
# config12 = {"Vliq1":340, "Vliq2":3400, "t_resx1":1.5, "t_resx2":700, "Q1":618, "Q2":618}
config12 = {"Vliq1":340, "Vliq2":3400, "t_resx1":1.5, "t_resx2":700, "Q1":618}
[15]:
result_config12 = adm1fu.reactor2(**config12)
Reactor run, phase-one:
$ADM1F_EXE -Vliq 340 -t_resx 1.5 -influent_file influent_cur.dat
indicator-051.out
Reactor run, phase-two:
$ADM1F_EXE -Vliq 3400 -t_resx 700 -influent_file influent_cur.dat
indicator-024.out
[16]:
# output using config12
result_config12 = adm1fu.reactor2(**config12)
ls1_config12 = result_config12[0].tolist()
ls2_config12 = result_config12[1].tolist()
df_config12 = pd.DataFrame(data = [ls1_config12],columns=output_name,index=['Phase1'])
df_config12.loc['Phase2'] = ls2_config12
df_config12
Reactor run, phase-one:
$ADM1F_EXE -Vliq 340 -t_resx 1.5 -influent_file influent_cur.dat
indicator-051.out
Reactor run, phase-two:
$ADM1F_EXE -Vliq 3400 -t_resx 700 -influent_file influent_cur.dat
indicator-024.out
[16]:
| Ssu | Saa | Sfa | Sva | Sbu | Spro | Sac | Sh2 | Sch4 | Sic | ... | Alk | NH3 | NH4 | LCFA | percentch4 | energych4 | efficiency | VFA/ALK | ACN | sampleT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phase1 | 80.561600 | 33.470400 | 7808.7500 | 2598.14000 | 4246.87000 | 3285.25000 | 9696.05 | 28.506900 | 7.771710e-32 | 147.214 | ... | 60316.4 | 0.048011 | 1145.6 | 7808.7500 | 3.068330e-32 | 104.203 | -104.737 | 0.308161 | 1.216580e-18 | 0.550162 |
| Phase2 | 0.355681 | 0.132504 | 19.2757 | 2.37729 | 3.05196 | 2.19399 | 1248.96 | 0.000046 | 7.114040e+01 | 867.356 | ... | 17101.8 | 15.289200 | 1139.4 | 19.2757 | 6.198330e+01 | 192.277 | -590.307 | 0.068884 | -2.807000e+01 | 6.241160 |
2 rows × 67 columns
[ ]: